
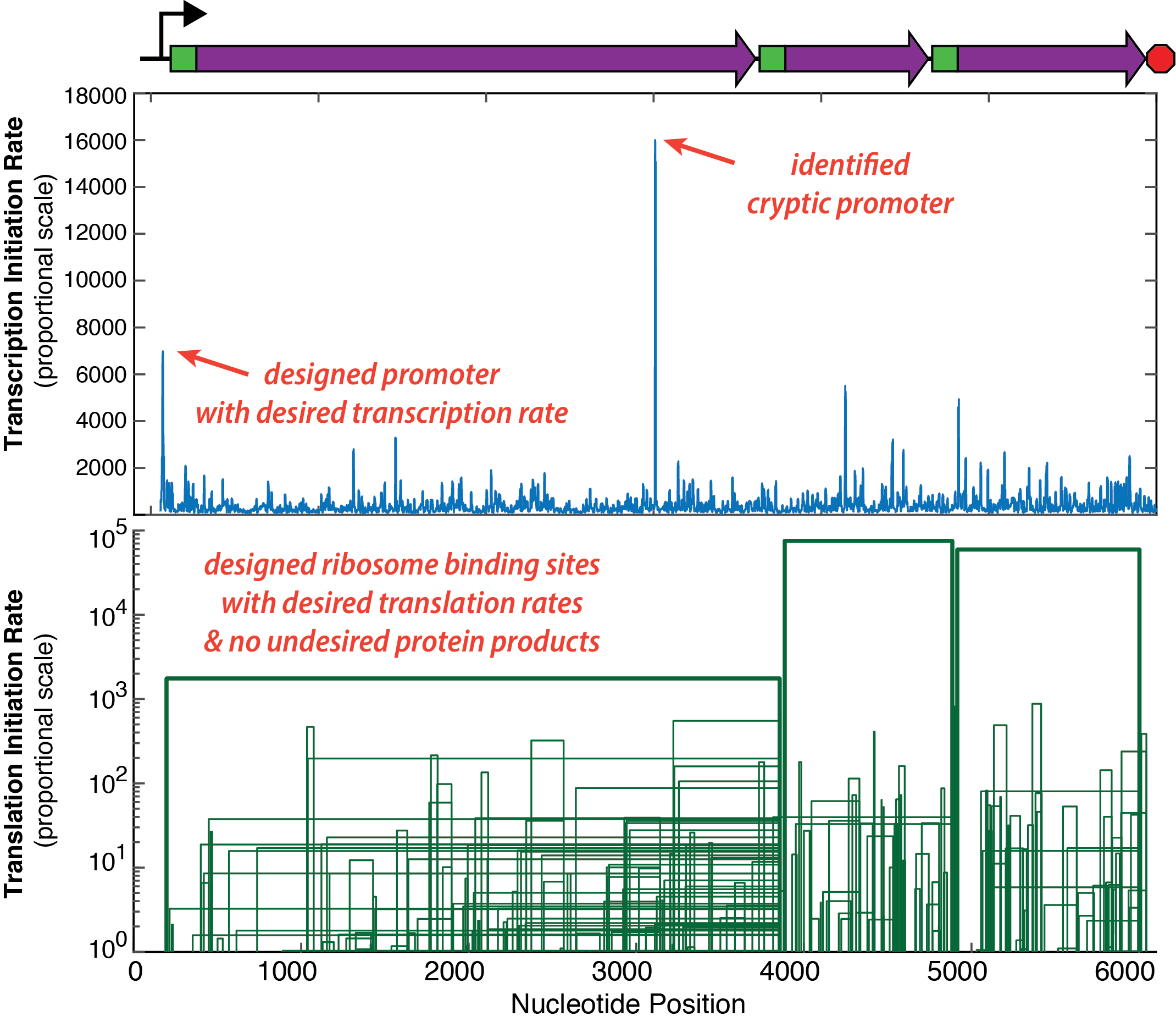
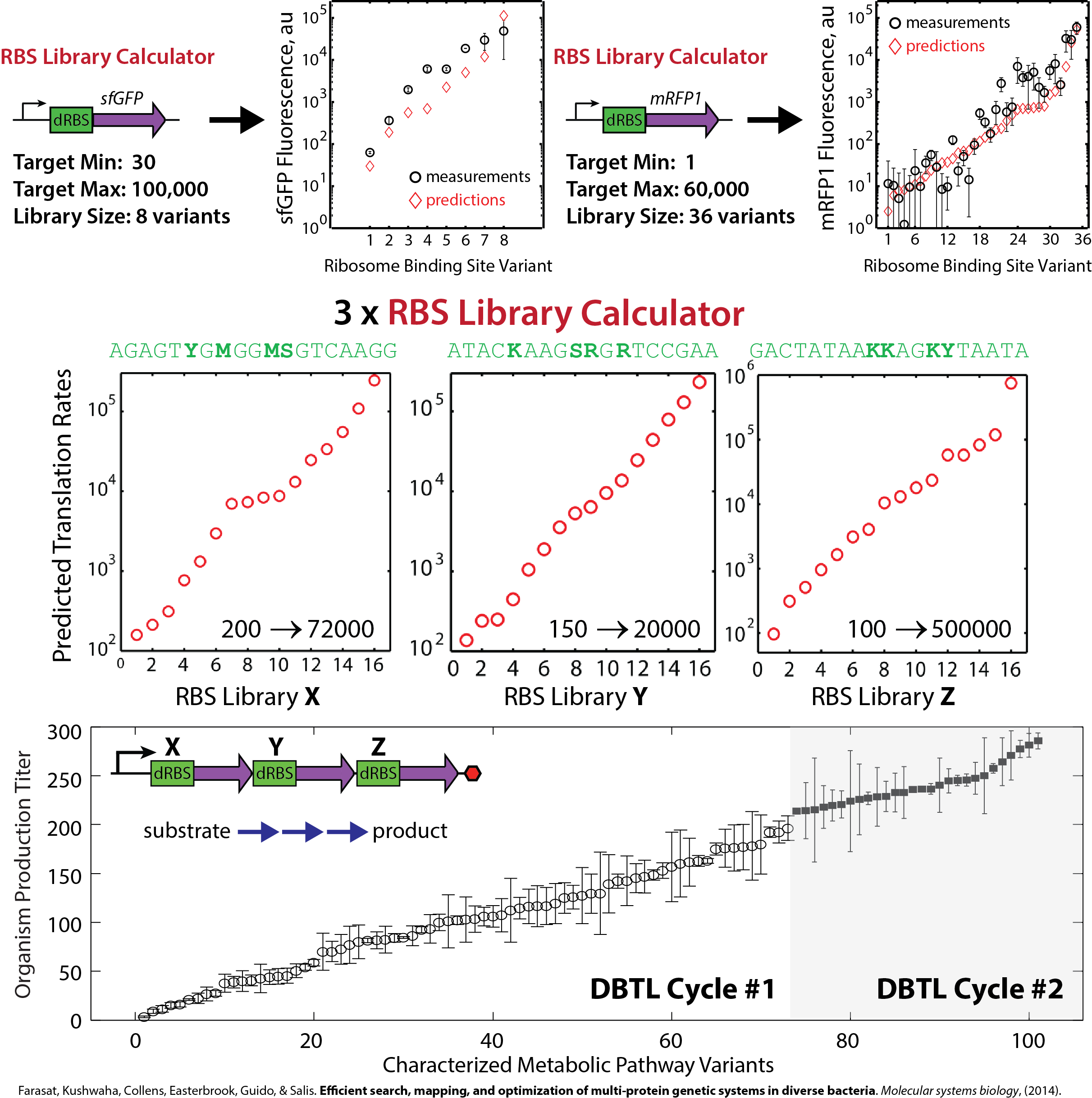
Calculate transcription rates, translation rates, and mRNA decay rates across your system. Find cryptic promoters and mis-annotated coding sequences. Design synthetic promoters and ribosome binding sites with desired protein expression levels.
Systematically vary and optimize enzyme expression levels. Find the rate-limiting steps. Redirect metabolic fluxes. Maximize organism productivities.
10 to 10000 Proteins. $100 per Cloned Expression Plasmid.
Any Amino Acid Sequence (up to 900 amino acids)
For bioprospecting, biologics development, enzyme engineering & much much more.

Research Scientist. Molecular Biology & Biochemistry.
Genomics. Next-gen Sequencing.

Chemical Engineer. Synthetic Biologist.
Software developer. Entrepreneur. Professor.

Researchers from MIT applied the RBS Library Calculator to systematically vary the expression of enzymes inside the nif gene cluster to maximize the rate of nitrogen fixation inside the soil microbe Rhizobium sp. IRBG74. They identified several rules to increase nitrogenase activity from engineered operons.
Researchers from MIT applied the RBS Calculator to optimize a genetic circuit in engineered L. lactis probiotic bacteria to detect and suppress cholera infection inside the body.
Researchers from the University of Manchester applied the RBS Library Calculator to systematically vary the expression of ten enzymes to maximize limonene production in E. coli, leveraging machine learning to identify optimal enzyme expression levels. Their approach was able to boost production titers by over 60% while screening less than 3% of the combinatorial RBS library.
Researchers from the University of Illinois at Urbana-Champaign applied the RBS Library Calculator to systematically vary enzyme expression levels to over-produce lycopene, utilizing a Bayesian optimization to optimally combine genetic parts and the iBioFab to automate the construction of desired strains.
Researchers from Washington University applied the RBS Calculator to vary the expression levels of two enzymes to maximize limonene production in Synechococcus 2973. They found that high Limonene Synthase expression and intermediate GPPS expression levels led to high limonene production titers.
Researchers from MIT utilized the RBS Calculator to optimize the expression of 12 different inducible transcription factors to create a Marionette E. coli strain, capable of tunably controlling the expression levels of 12 proteins using small molecule inducers.
Researchers from Columbia University applied the RBS Calculator to engineer a CRISPR-based genetic circuit that converts temporal biological signals into physical genomic modifications (CRISPR spacers), creating a biological tape recording of prior chemical events.
Researchers from the Joint BioEnergy Institute applied the RBS Calculator to vary and optimize enzyme expression levels to over-produce three types of C5 alcohols and to reduce the formation of toxic byproducts, achieving 70% of the maximum possible production titer.
Researchers from MIT applied the RBS Library Calculator to systematically vary the expression of 16 enzymes in the nif gene cluster, responsible for nitrogen fixation. They identified design rules and optimal expression levels to maximize nitrogenase activity.
Researchers from MIT applied the RBS Calculator to engineer genetic circuits that convert analog chemical signals into discrete cellular states with programmed Boolean logic, utilizing recombinases to re-orient regulatory genetic parts in response to desired signals.
Researchers from Duke University utilized the RBS Calculator to engineer a lysis-inducing genetic circuit to demonstrate that a programmed level of bacterial suicide actually improves population-level growth rates in response to stress, including antibiotic treatment.
Researchers from Harvard University employed the RBS Calculator to design RNA scaffolds that co-position multiple enzymes inside cells to accelerate their reaction rates, selecting RNA sequences that have the lowest possible translation initiation rates to prevent the ribosome from disrupting scaffold assembly.
Researchers from the Universidad Nacional Autónoma de México utilized the RBS Calculator to control the expression of five enzymes to over-produce resveratrol, using a co-culture of two engineered E. coli strains to share the production workload.
Researchers from the University of Wisconsin-Madison applied the RBS Library Calculator to systematically vary the expression of five enzymes responsible for isobutanol production in E. coli. From these results, they identified rate-limiting steps and achieved high isobutanol production titers.
Researchers from the Joint Bioenergy Institute and Lawrence Berkeley National Lab applied the RBS Library Calculator to systematically vary the expression of the Crp transcription factor inside E. coli to maximally over-produce methyl ketone. When utilizing a feedstock containing both glucose and xylose, they found that reducing Crp expression was necessary to achieve high methyl ketone production titers.
Researchers from Penn State University combined the Operon Calculator and the RBS Library Calculator to design and optimize a 5-enzyme Entner–Doudoroff pathway that regenerates NADPH. They identified the optimal enzyme expression levels that increased NADPH regeneration by over 25-fold.
Researchers from MIT and NIST utilized the RBS Calculator to tune the expression of transcription factors to develop modular genetic circuits with desired Boolean programs, utilizing automated design to combine genetic circuit modules.
Researchers from the University of Queensland, Brisbane applied the Operon Calculator and the RBS Calculator to express the multi-enzyme Wood-Werkman cycle in E. coli in order to over-produce propionic acid. Systematic optimization yielded a 5-fold increase in proprionic acid production titers.
Researchers from the University of Colorado Boulder combined the RBS Library Calculator with multiplexed genome recombineering to systematically vary the expression of enzymes responsible for hydrolysate and isobutanol tolerance. Using a novel sequencing approach (TRACE), they identified enzyme expression levels that maximized the organism's growth rate at high concentrations of hydrolyzed lignocellulose and high concentrations of isobutanol.
Researchers from MIT applied the RBS Calculator to optimize the expression of five antimicrobial peptides, delivered using phagemids that target pathogenic bacteria inside the human body.
Researchers from Texas A&M University applied the RBS Calculator to increase expression of enzymes for higher bioconversion of CO2 and light to limonene in Synechococcus elongatus, increasing limonene production titers by 100-fold.
Researchers from A*STAR Singapore utilized the RBS Calculator to increase the expression of two enzymes responsible for viridiflorol production in E. coli.
Researchers from MIT applied the RBS Library Calculator to systematically vary and optimize the expression levels of TetR-homolog transcription factors to develop a toolbox of high-performance genetic circuits that can be used to program cellular behaviors.
Researchers from the National Renewable Energy Laboratory and the University of Colorado Boulder utilized the RBS Library Calculator to systematically optimize a multi-enzyme pathway to over-produce styrene in E. coli. They found that some enzymes must be expressed at intermediate levels to achieve maximal production titers.
Researchers from Jiangnan University applied the RBS Calculator to evaluate how changing the expression level of leech hyaluronidase enzyme varied the production rate and molecular weight of hyaluronan polymers, using Bacillus subtilis as the production organism.
Researchers from Rice University utilized the RBS Calculator to control expression of light-sensing optogenetic circuit components, enabling rapid temporal control over gene expression levels and genetic circuit functions using two different wavelengths of light.
Researchers from the University of Michigan utilized the RBS Calculator to understand the effects of sequence mutations gained during adaptive laboratory evolution to achieve tolerance at higher isobutanol concentrations.
Researchers from MIT utilized the RBS Library Calculator to optimize the expression of DNA-flipping integrases, enabling the permanent storage and utilization of digital information inside a genetic circuit.
Researchers from the University of Colorado Boulder utilized the Operon Calculator and the RBS Library Calculator to design a synthetic operon that expresses 7 urease enzymes responsible for CaCO3 production in E. coli. By varying enzyme expression levels, they found an inverse relationship between urease expression levels and CaCO3 polycrystal size.
Researchers at the Tianjin Institute of Industrial Biotechnology and the Dalian Polytechnic University applied the RBS Library Calculator to systematically vary both genomic and plasmid-based enzyme expression levels to over-produce hydrogenobyrinic acid. They found that fine-tuning the expression of endogenous enzymes was necessary to minimize the production of toxic intermediate compounds and maximize the production of hydrogenobyrinic acid.